O-GlcNAcylation functions in a protein- and site-specific manner. Despite the great progress in experimentally mapping O-GlcNAc sites, it is still a challenging task in many cases. We constructed an ensemble model O-GlcNAcPRED-DL to predict O-GlcNAc sites on proteins by using deep learning approaches. In comparison to existing methods, O-GlcNAcPRED-DL shows improved sensitivity and accuracy. We anticipate that it will be used as a valuable tool to expedite the discovery of O-GlcNAc sites on proteins (especially of humans and mice) and elucidation of their functions in physiology and diseases.
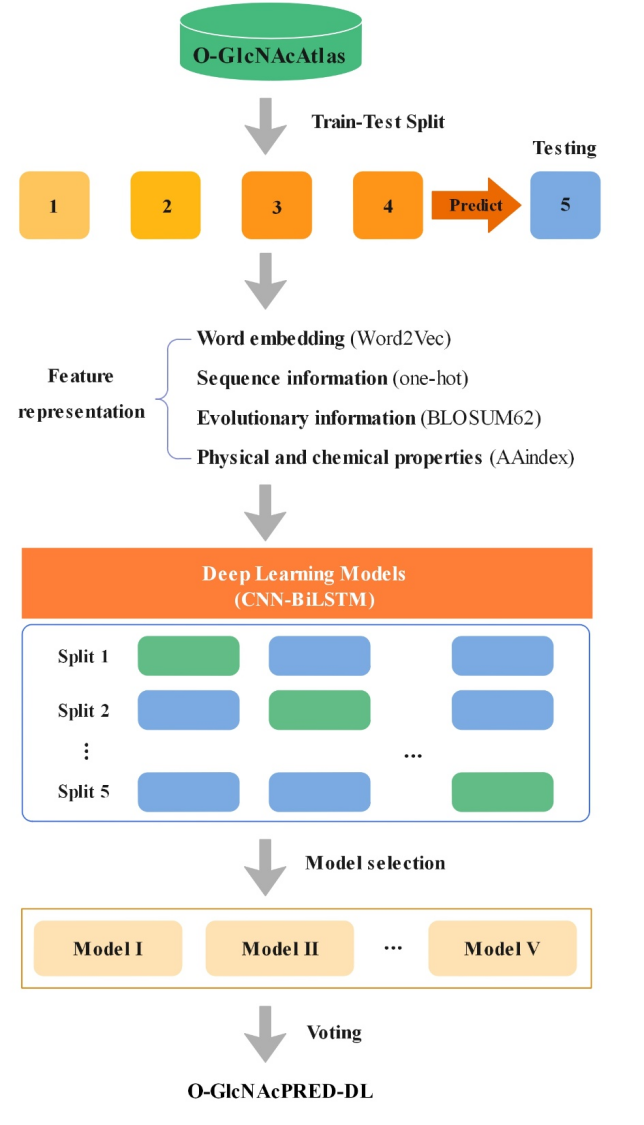
If you find this resource helpful, please cite our paper (Fengzhu Hu,# Weiyu Li,# Yaoxiang Li, Chunyan Hou, Junfeng Ma,* Cangzhi Jia.* O-GlcNAcPRED-DL: prediction of protein O-GlcNAcylation sites based on an ensemble model of deep learning. Journal of Proteome Research. 2024, 23, 1, 95–106. https://pubs.acs.org/doi/10.1021/acs.jproteome.3c00458)